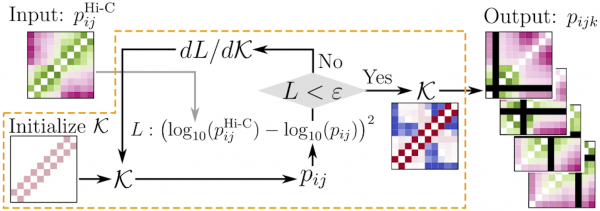
Extracting multi-way chromatin contacts from Hi-C data
2021
期刊
PLOS Computational Biology
下载全文
- 卷 17
- 期 12
- 页码 e1009669
- Public Library of Science (PLoS)
- ISSN: 1553-7358
- DOI: 10.1371/journal.pcbi.1009669
